Diabetes Data and Hypothesis Hub (D2H2)
A platform that facilitates data-driven hypothesis generation for the diabetes and related metabolic disorder research community.


ICE status:
Peer connection status:
Signaling status:
Streaming status:


Examples
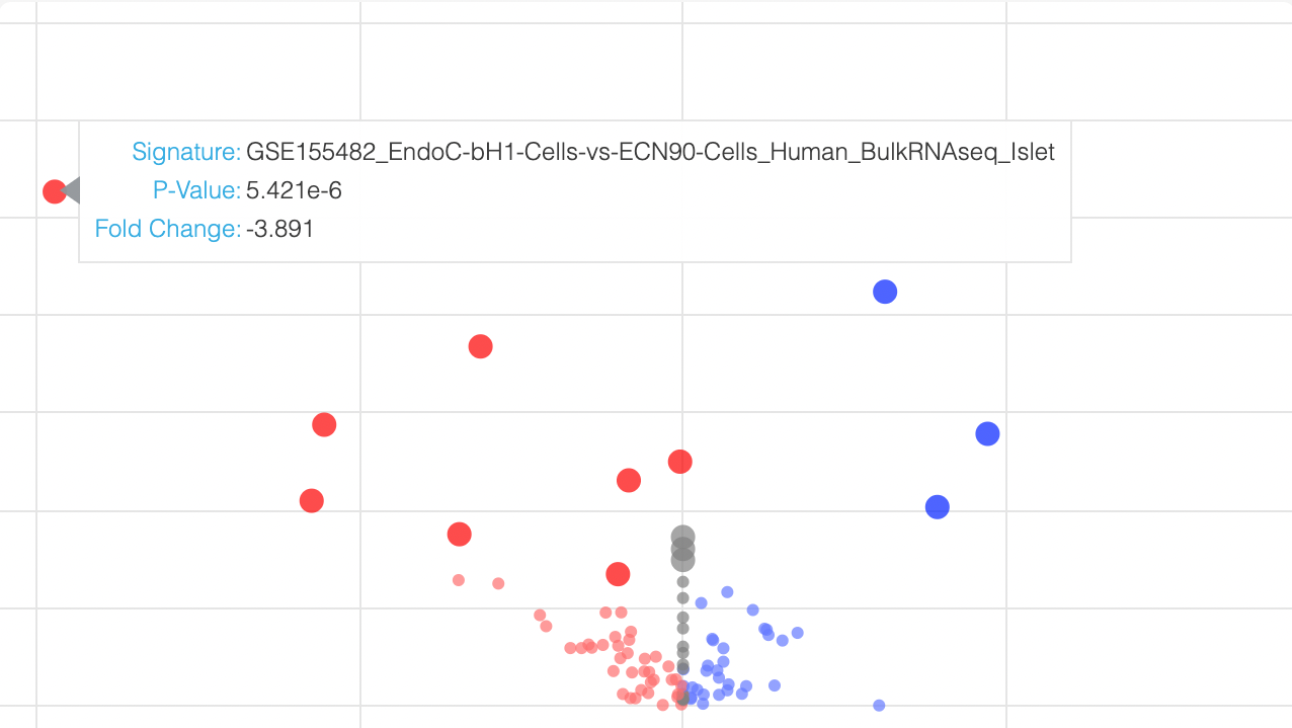
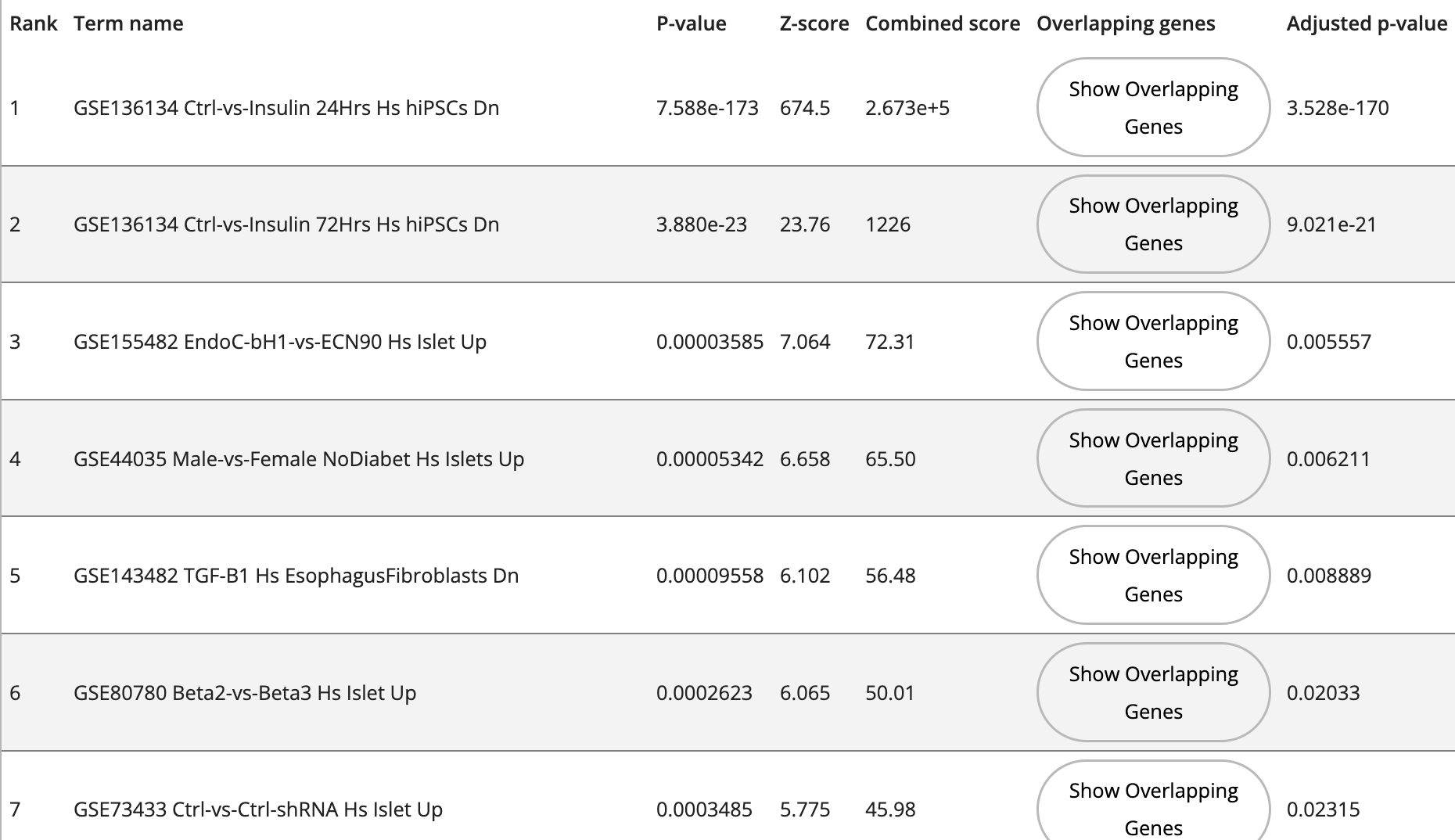
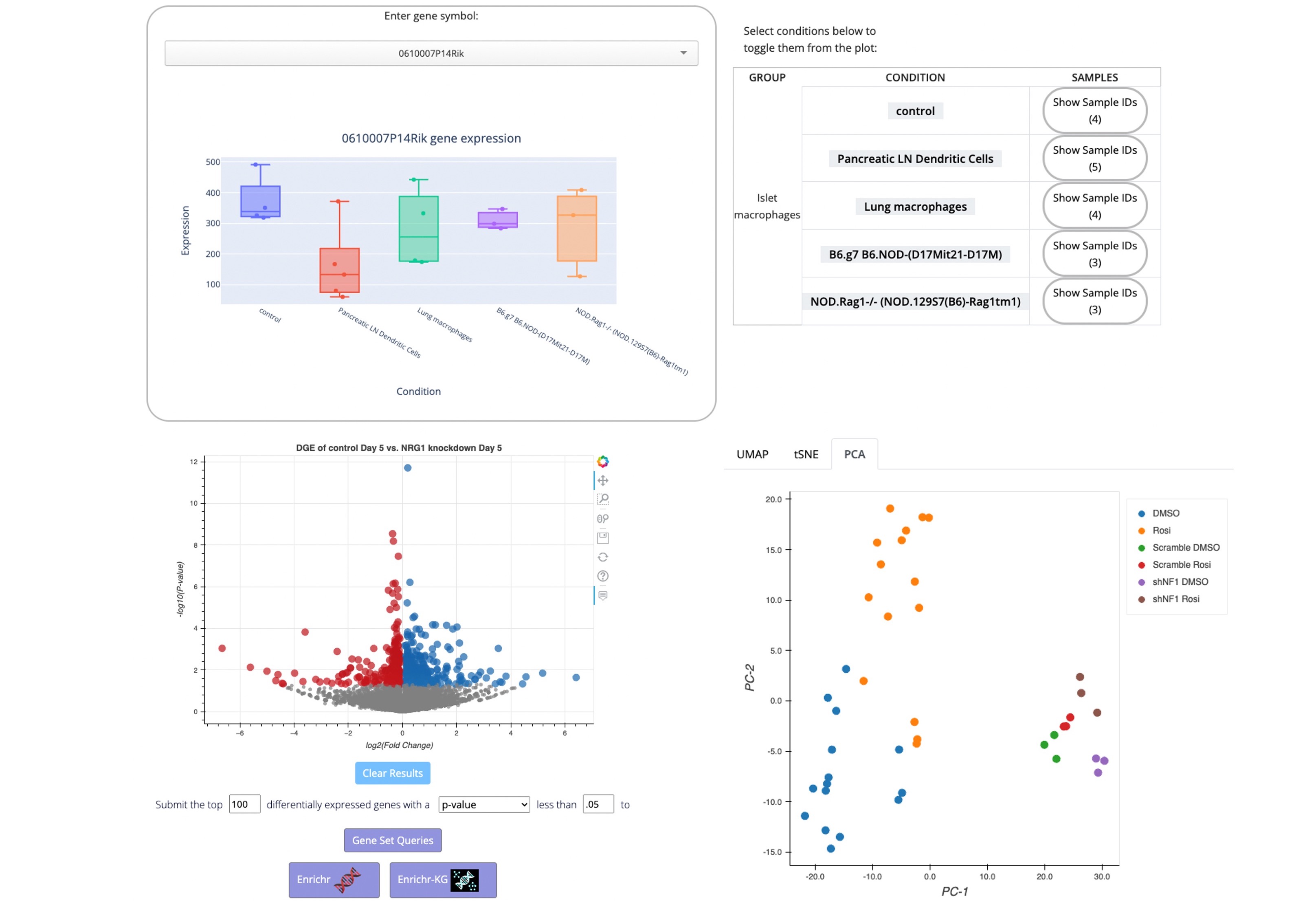
Explore a collection of curated diabetes related Bulk-RNA-seq and microarray studies manually extracted and processed from NCBI's GEO. The expression level of single genes across conditions is visualized as a customized box plot viewer; the samples can be visualized as UMAP, tSNE and PCA plots; and users cam compute differential expression for any two conditions with replicates.
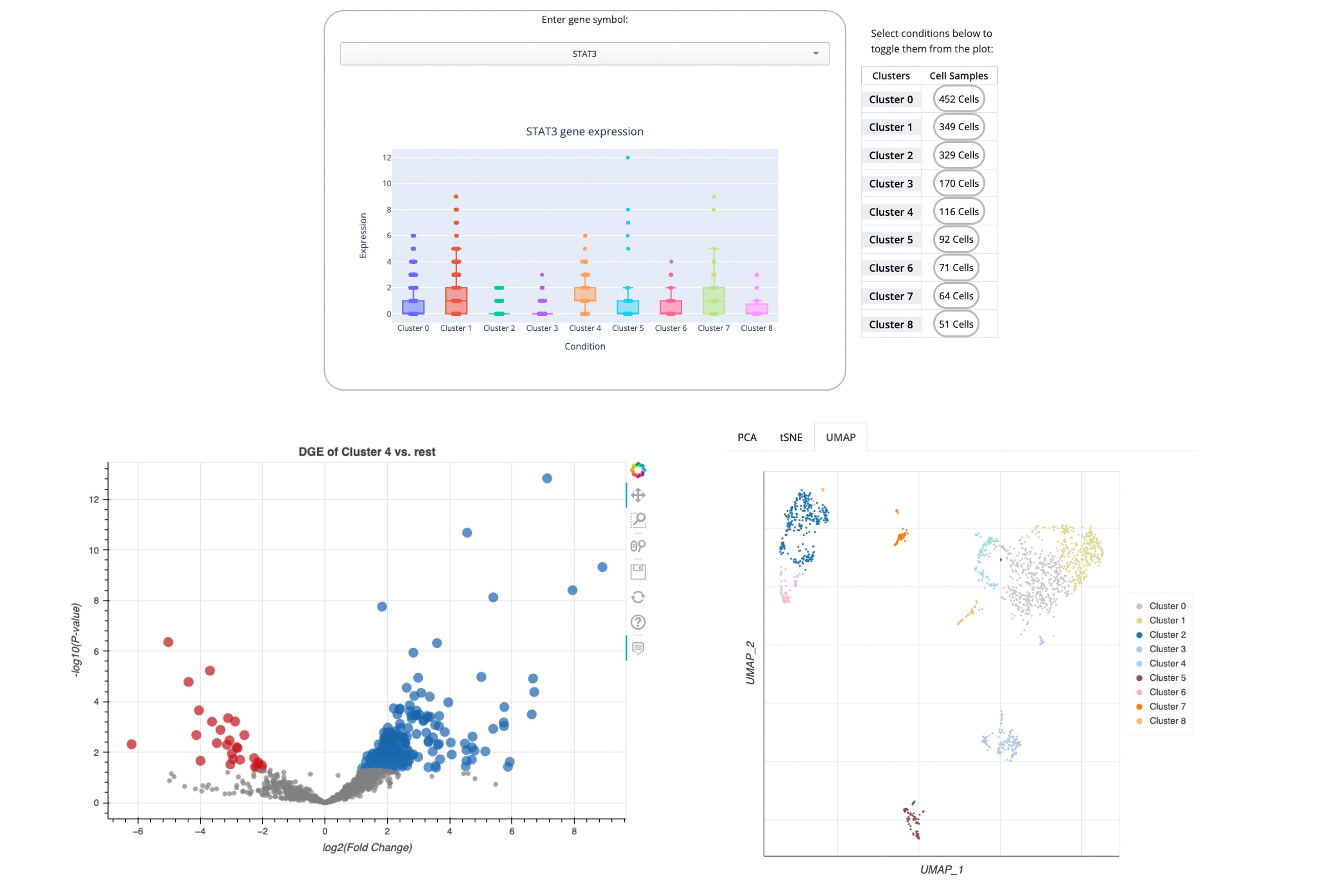
Explore a collection of curated diabetes related scRNA-seq studies extracted from NCBI's GEO. The expression of single genes across clusters of a chosen profile can be visualized as a customized box plot viewer; the single cells can be visualized in a UMAP, t-SNE and PCA; and users can compute differential expression for each automatically identified cluster
 Chatbot
Chatbot Single Gene Queries
Single Gene Queries
 Gene Set Queries
Gene Set Queries
 Bulk Studies
Bulk Studies
 Single Cell Studies
Single Cell Studies
 Hypotheses
Hypotheses
 Resources
Resources
 Contribute
Contribute
 Downloads
Downloads About
About
 Help
Help